Different Sponge Species Have Highly Specific and Stable Microbiomes, MBL Scientists Report

Contact: Diana Kenney
508-2897139; dkenney@mbl.edu
WOODS HOLE, Mass. —The sea sponge is about as simple as an animal can get, but its associated bacterial community—its microbiome —is known to approach the complexity of the diverse microbiome in the human gut.
Now, scientists at the Marine Biological Laboratory (MBL) have shown that different species of Hexadella sponges each have a highly specific and stable microbiome, not only in terms of the most abundant members of the associated microbial community, but the rare members as well. They published their results recently in the International Society for Microbial Ecology (ISME) Journal.
“When we looked at what microbial community occurred in a species of sponge, we always found the same community, no matter where geographically and at which depth the sponge [lived],” says lead author Julie Reveillaud, a postdoctoral associate in the MBL’s Bay Paul Center.
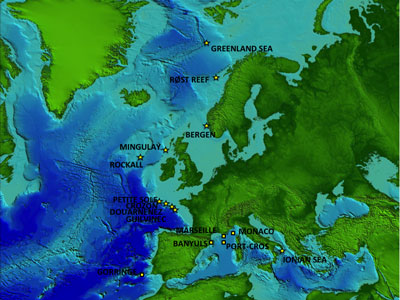 Map displaying Hexadella sampling locations: deep-sea sites (star shape) and shallow-water sites (square shape). Map provided by the project Hotspot Ecosystems Research and Man’s Impact on European Seas (HERMOINE).
Map displaying Hexadella sampling locations: deep-sea sites (star shape) and shallow-water sites (square shape). Map provided by the project Hotspot Ecosystems Research and Man’s Impact on European Seas (HERMOINE).Reveillaud’s team was interested in whether closely related sponge species display a higher degree of similarity in the composition of their microbiomes than do more distantly related sponge species. They found a weak correlation between the sponges’ relatedness and their microbiomes, indicating Hexadella species did not inherit their entire microbiomes from their parents (vertical transmission), but also acquired microbes horizontally from the environment.
What’s interesting, Reveillaud says, is while both horizontal and vertical transmission of microbes are occurring, the microbiomes were found to be highly specific to their sponge host species. That indicates that the microbes acquired horizontally are not just randomly selected from the environment and maintained. “There is a mechanism by which the sponge and the microbiome are specifically selecting each other,” Reveillaud says.
 Hexadella dedritifera (yellow) being sampled with the ROV Victor in the deep-sea reefs in Europe. Photo credit: Ifremer, France.
Hexadella dedritifera (yellow) being sampled with the ROV Victor in the deep-sea reefs in Europe. Photo credit: Ifremer, France.Using “ultra-deep” DNA sequencing technology to obtain very fine-grained pictures of the microbiomes’ compositions, the team compared seven sponge species from the genus Hexadella whose habitats range from the Mediterranean Sea to the Greenland Sea, and who live at depths ranging from 15 to 1,000 meters. It was exciting to discover the high degree to which the microbiomes are specific to their hosts across such a wide geographic distribution, Reveillaud says.
Several other scientists in the Bay Paul Center collaborated on the study, including Loïs Maignien, A. Murat Eren, Julie Huber, and Mitchell Sogin.
Citation:
Reveillaud J, Maignien L, Eren AM, Huber JA, Apprill A, Sogin ML and Vanreusel A (2014) Host-specificity among abundant and rare taxa in the sponge microbiome. ISME Journal, advance online publication January 9; doi: 10.1038/ismej.2013.227.
—###—
The Marine Biological Laboratory (MBL) is dedicated to scientific discovery and improving the human condition through research and education in biology, biomedicine, and environmental science. Founded in Woods Hole, Massachusetts, in 1888, the MBL is a private, nonprofit institution and an affiliate of the University of Chicago.