MBL Awards Whitman Center Fellowships to 27 Exceptional Investigators

WOODS HOLE, Mass. – A diverse group of 27 scientists from 24 universities and research institutes worldwide have been named 2019 Whitman Center Fellows by the Marine Biological Laboratory (MBL). These fellowships enable exceptional scientists, including Early Career Fellows, to conduct independent research at the MBL and take advantage of its unique resources and highly collaborative scientific community.
The Whitman Center Fellows will be supported for up to 10 weeks to pursue research, particularly within these fields of strategic vision for MBL:
- Evolutionary, genetic, and genomic approaches in regenerative biology, developmental biology, and neuroscience, with an emphasis on novel marine organisms
- Integrated imaging and computational approaches to illuminate cellular function and biology
- Integrated approaches to the study of microbial communities in coastal environments
“This year’s Whitman Fellows bring a wealth of scientific talent to the MBL’s convening environment, which is dedicated to offering scientists the space, time, inspiration, and support to intensively pursue research of fundamental importance,” said David Mark Welch, Director of the MBL Division of Research.
The MBL provides access to state-of-the-art instrumentation, innovative imaging technology and research, genome sequencing, model marine and freshwater organisms, and modern laboratory facilities. It also offers an exceptionally dynamic, interactive, and creative scientific environment. In addition to its resident research programs, the MBL annually convenes hundreds of principal investigators, postdocs, graduate students, and research associates from around the world to participate in Whitman Center research, scientific discovery courses, lectures, and field studies.
Several of the 2019 Whitman Fellows are coming to the MBL for the first time to launch a new project, while others will continue research programs they established in the Whitman Center in prior years. Many of the organisms the fellows are studying at MBL are pictured on this page (captions below). The fellows are:
Whitman Center Early Career Fellows
Amy Herbert, Stanford University
Sea robins as a model for the genetic basis of novel traits in vertebrates
Hooi Lynn Kee, Stetson University
Investigating the microbial rainbow of iridescent marine bacteria
Matthew McCoy, Stanford University
The role of gene complexity in generating neuronal diversity within cephalopod nervous systems
Rachel Miller, The University of Texas Medical School at Houston
Strategies to advance the study of kidney development in Xenopus
Sy Redding, University of California, San Francisco
Is protein aggregation important biology? Measuring the dynamic properties of protein assemblies on DNA and chromatin
Stefanie Redemann, University of Virginia
Studying spindle mechanics by light microscopy, electron tomography, biophysical modeling and simulation
Shashank Shekhar, Brandeis University
Unraveling the physical origins of multicellularity using a novel model organism
Oleg Simakov, University of Vienna, Austria
Are there gene regulatory networks that are specific for cephalopod novelties?
Victoria Sleight, University of Cambridge, UK
Molluscan biomineralization: development, repair and evolution
Trevor Wardill, University of Minnesota
Predictive control of eye movements and strike accuracy in the cuttlefish brain
Shigeki Watanabe, Johns Hopkins University
Molecular mechanisms of synaptic transmission in zebrafish photoreceptors
Elizabeth Wilbanks, University of California, Santa Barbara
Microbiomes from genotype to phenotype: Assessing the effect of strain-level bacterial diversification on cryptic sulfur cycling in the pink berry microbial consortia
Christina Zakas, New York University
Establishing transgenic lines in Streblospio benedicti, a new model for evolutionary developmental biology
Whitman Center Fellows
Tobias Baskin, University of Massachusetts Amherst
Role of microtubules in controlling cellulose organization in plant cells
Lionel Christiaen, New York University
Novel functional genomics tools for the chordate model Ciona
Karen Crawford, St. Mary's College of Maryland
Optimizing genome editing methods for Doryteuthis peallii: Building upon the successful generation of the first CRISPR-Cas9 gene knock outs from summer 2018
André Fenton, New York University
Starting to integrate across levels of biology to understand how silencing a gene causes intellectual disability and autism
Ben Evans, McMaster University, Canada
Developmental systems drift of sex determination in African clawed frogs (Xenopus)
Uri Gat, Hebrew University in Jerusalem, Israel
A comparative study between two highly regenerative aquatic model animals: The function of Cthrc genes in the regeneration programs of the salamander Axolotl and the sea anemone Nematostella
Edgar Gomes, Instituto Medicina Molecular, Portugal
Moving the cell nucleus in migrating cells
Jeffrey Gross, University of Pittsburgh School of Medicine
Regeneration of the zebrafish retinal pigment epithelium
Phillippe Laissue, University of Essex, UK
Developing models for integral studies of coral bleaching and recovery
Paul Maddox, University of North Carolina, Chapel Hill
LITE imaging of cell division
Isa Schön, Royal Belgian Institute of Natural Sciences, Belgium
Developing ostracods as emerging aquatic model organisms for evolutionary and biodiversity research
Ava Udvadia, University of Wisconsin, Milwaukee
Elucidating evolutionarily conserved mechanisms regulating successful optic nerve regeneration in vertebrate species
Denisa Wagner, Boston Children’s Hospital
Delineation of the role of the NLRP3 inflammasome in NETosis by time-lapse microscopy
Rafael Yuste, Columbia University
HydraLab 2019: Experimental and Computational Methods for Neural Circuit Analysis
Homepage photo: Amy Herbert of Stanford University observing sea robin development. Credit: Matt McCoy. (Herbert and McCoy were Grass Fellows at MBL in 2018 and are among this year's Whitman Early Career Fellows).
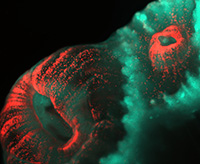
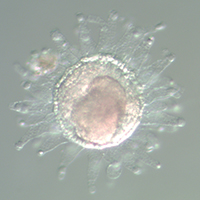
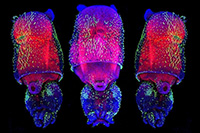
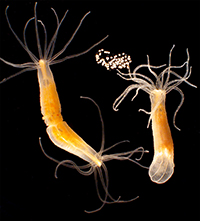
Among the organisms that the 2019 Whitman Fellows are studying are, from top, left to right: Coral (Astrangia poculata) (Credit: Phillipe Laissue); Sea squirt (Ciona intestinalis) (MBL Logan Science Journalism Program); Squid (Doryteuthis pealeii) (Wang Chi Lau, MBL Embryology Course); Starlet sea anemone (Nematostella vectensis) (John Finnerty); Axolotl (Ambystoma mexicanum) (Dee Sullivan); Zebrafish (Danio rerio) (IMP Vienna); Sea robin (Triglidae) (Doug McFarlane); African clawed frog (Xenopus laevis) (National Xenopus Resource); Seed shrimp (Ostracoda) (Anna Syme); Cuttlefish (Sepia bandensis) (Tom Kleindinst); Pink berry microbial consortia (Elizabeth Wilbanks); Bar-gilled mud worm (Streblospio benedicti) (E. Lazo-Wasem); Tobacco plant (Nicotiana tabacum) (Jon Sullivan); Hydra (Stefan Seibert); Iridescent marine bacteria (Sarah Lawhun); Slipper limpet (Crepidula fornicata) (E. Lazo-Wasem).