Team Dramatically Reduces Image Analysis Times Using Deep Learning, Other Approaches

Media Contact: dkenney@mbl.edu; 508-685-3525
WOODS HOLE, Mass. – A picture is worth a thousand words –but only when it’s clear what it depicts. And therein lies the rub in making images or videos of microscopic life. While modern microscopes can generate huge amounts of image data from living tissues or cells within a few seconds, extracting meaningful biological information from that data can take hours or even weeks of laborious analysis.
To loosen this major bottleneck, a team led by MBL Fellow Hari Shroff has devised deep-learning and other computational approaches that dramatically reduce image-analysis time by orders of magnitude -- in some cases, matching the speed of data acquisition itself. They report their results this week in Nature Biotechnology.
“It’s like drinking from a firehose without being able to digest what you’re drinking,” says Shroff of the common problem of having too much imaging data and not enough post-processing power. The team’s improvements, which stem from an ongoing collaboration at the Marine Biological Laboratory (MBL), speed up image analysis in three major ways.
First, imaging data off the microscope is typically corrupted by blurring. To lessen the blur, an iterative “deconvolution” process is used. The computer goes back and forth between the blurred image and an estimate of the actual object, until it reaches convergence on a best estimate of the real thing.
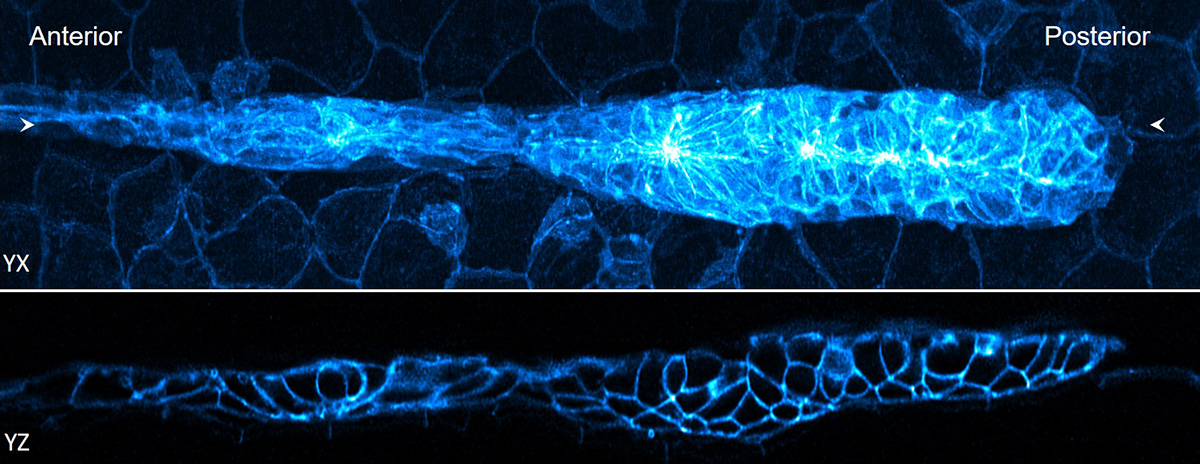 Lateral and axial images of 32-hour zebrafish embryo, marking cell boundaries within and outside the lateral line primordium. Credit: H. Vishwasrao and D. Dalle Nogare, National Institutes of Health
Lateral and axial images of 32-hour zebrafish embryo, marking cell boundaries within and outside the lateral line primordium. Credit: H. Vishwasrao and D. Dalle Nogare, National Institutes of HealthBy tinkering with the classic algorithm for deconvolution, Shroff and co-authors accelerated deconvolution by more than 10-fold. Their improved algorithm is widely applicable “to almost any fluorescence microscope,” Shroff says. “It’s a strict win, we think. We’ve released the code and other groups are already using it.”
Next, they addressed the problem of 3D registration: aligning and fusing multiple images of an object taken from different angles. “It turns out that it takes much longer to register large datasets, like for light-sheet microscopy, than it does to deconvolve them,” Shroff says. They found several ways to accelerate 3D registration, including moving it to the computer’s graphics processing unit (GPU). This gave them a 10- to more than 100-fold improvement in processing speed over using the computer’s central processing unit (CPU).
“Our improvements in registration and deconvolution mean that for datasets that fit onto a graphics card, image analysis can in principle keep up with the speed of acquisition,” Shroff says. “For bigger datasets, we found a way to efficiently carve them up into chunks, pass each chunk to the GPU, do the registration and deconvolution, and then stitch those pieces back together. That’s very important if you want to image large pieces of tissue, for example, from a marine animal, or if you are clearing an organ to make it transparent to put on the microscope. Some forms of large microscopy are really enabled and sped up by these two advances.”
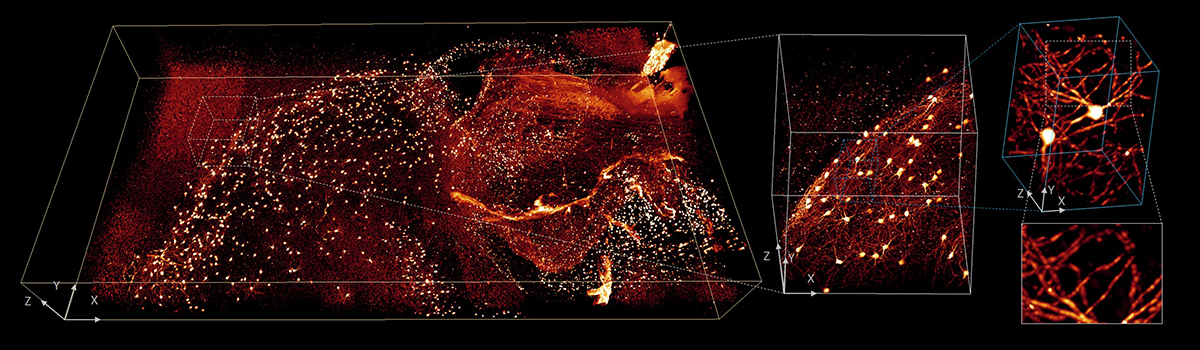 4 x 2 x 0.5 mm3 volume of brain from fixed and cleared mouse. Progressively higher resolution subvolumes highlight the detailed structures with isotropic submicron resolution. Credit: Y. Su, Y. Wu, H. Vishwasrao and T. Usdin, National Institutes of Health
4 x 2 x 0.5 mm3 volume of brain from fixed and cleared mouse. Progressively higher resolution subvolumes highlight the detailed structures with isotropic submicron resolution. Credit: Y. Su, Y. Wu, H. Vishwasrao and T. Usdin, National Institutes of HealthLastly, the team used deep learning to accelerate “complex deconvolution” – intractable datasets in which the blur varies significantly in different parts of the image. They trained the computer to recognize the relationship between badly blurred data (the input) and a cleaned, deconvolved image (the output). Then they gave it blurred data it hadn’t seen before. “It worked really well; the trained neural network could produce deconvolved results really fast,” Shroff says. “That’s where we got thousands-fold improvements in deconvolution speed.”
While the deep learning algorithms worked surprisingly well, “it’s with the caveat that they are brittle,” Shroff says. “Meaning, once you’ve trained the neural network to recognize a type of image, say a cell with mitochondria, it will deconvolve those images very well. But if you give it an image that is a bit different, say the cell’s plasma membrane, it produces artifacts. It’s easy to fool the neural network.” An active area of research is creating neural networks that work in a more generalized way.
“Deep learning augments what is possible,” Shroff says. “It’s a good tool for analyzing datasets that would be difficult any other way.”
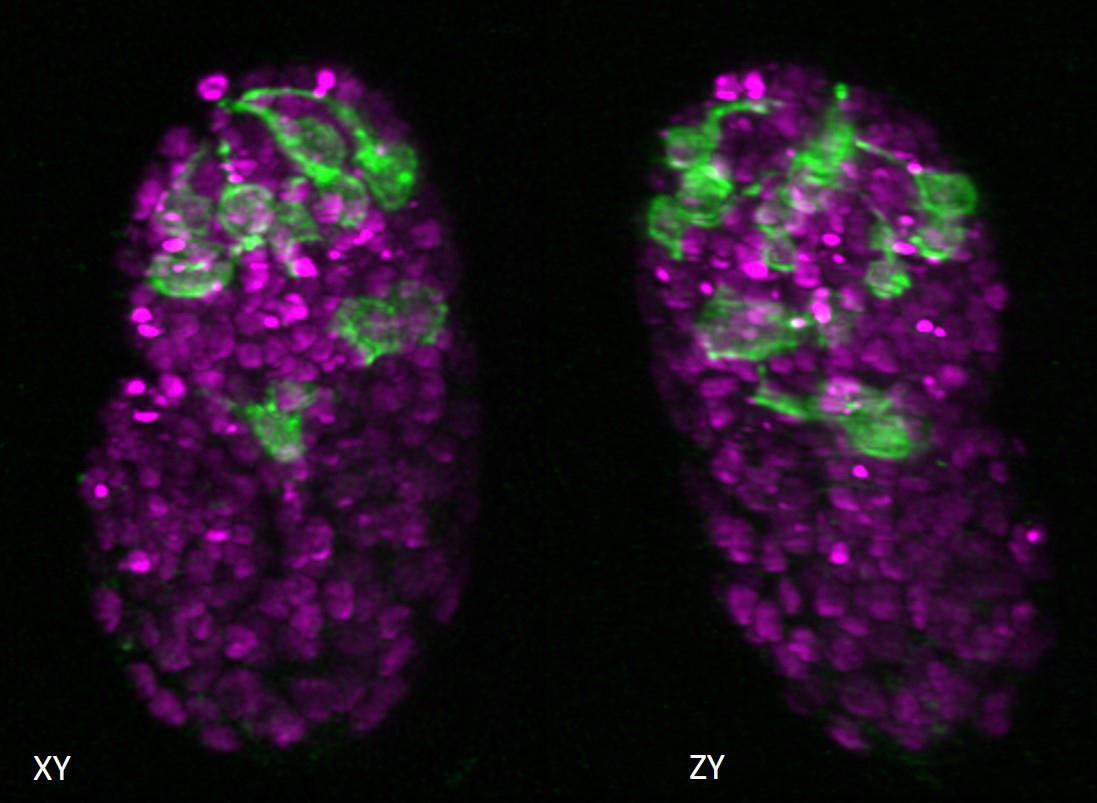 Lateral and axial images of C. elegans embryo expressing neuronal (green) and pan-nuclear (magenta) markers. Isotropic resolution enables lineage tracing and inspection of neurite outgrowth pre-twitching. Credit: L. Duncan and M. Moyle, Yale School of Medicine
Lateral and axial images of C. elegans embryo expressing neuronal (green) and pan-nuclear (magenta) markers. Isotropic resolution enables lineage tracing and inspection of neurite outgrowth pre-twitching. Credit: L. Duncan and M. Moyle, Yale School of MedicineThis research stems from an ongoing collaboration in the MBL Whitman Center between MBL Fellows Hari Shroff, senior investigator at the National Institute of Biomedical Imaging and Bioengineering; Patrick La Rivière, professor at the University of Chicago; and Daniel Colón-Ramos, professor at Yale Medical School. First author Min Guo is postdoctoral fellow in Shroff’s lab and a former teaching assistant for Shroff in the MBL’s Optical Microscopy and Imaging in the Biological Sciences course.
Citation: Min Guo et al (2020) Rapid image deconvolution and multiview fusion for optical microscopy. Nature Biotechnology: DOI: 10.1038/s41587-020-0560-x
Struggling with large imaging datasets is a common problem for biologists. “You need sophisticated computational capacity to use the powerful optical microscopes we have and are building,” says Abhishek Kumar, MBL Investigator and CZI Scientist. “Not all labs have the infrastructure required. And not all biologists have a PhD in computer science or mathematics!”
To address this need, the MBL is opening an Image Analysis Laboratory in the Lillie Building. The lab will be equipped with high-end, user-friendly workstations so biologists can bring in their imaging data and learn to analyze it themselves.
“We are designing the lab so people without a lot of computational expertise can use these technologies,” Kumar says.
The lab is part of the MBL’s Imaging Initiative, which convenes biologists, instrument developers, and computational scientists to advance research and training in next-generation microscopy and computational image analysis. Microscope developer Hari Shroff and paper co-authors Patrick La Rivière (a computational scientist) and Daniel Colón-Ramos (a neurobiologist) are among the collaborators in the Imaging Initiative.
“We are presented with a wide range of biological samples and biological problems at the MBL,” Kumar says. “And there is no universal microscope. No one instrument can answer all questions. Our goals in the Imaging Initiative are to develop instruments to address many different biological problems; improve image analysis capabilities, which are a bottleneck; and provide imaging technologies and solutions that are user-friendly, so a wide range of students and scientists can adopt them for their research.”
The MBL Image Analysis lab will be available to MBL students, Whitman Center scientists, and visitors in 2021.
—###—
The Marine Biological Laboratory (MBL) is dedicated to scientific discovery – exploring fundamental biology, understanding marine biodiversity and the environment, and informing the human condition through research and education. Founded in Woods Hole, Massachusetts in 1888, the MBL is a private, nonprofit institution and an affiliate of the University of Chicago.